CRISPR/Cas & co
- Date The exact date of the course will be announced when enough students have registered. You will then be contacted to confirm your availability
- Duration 4 days – 20 hours Doctoral School equivalent
- Language English and/or French (depending on participants; all supportive documents will be provided in English)
- Name of the teachers Johannes STUTTMANN, Sylvain FOCHESATO, Florian VEILLET, Pascaline AUROY-TARRAGO
- Administrative head Mila Sirinelli
- Number of students Group of 4-8 students
- Location CEA Luminy
Description:
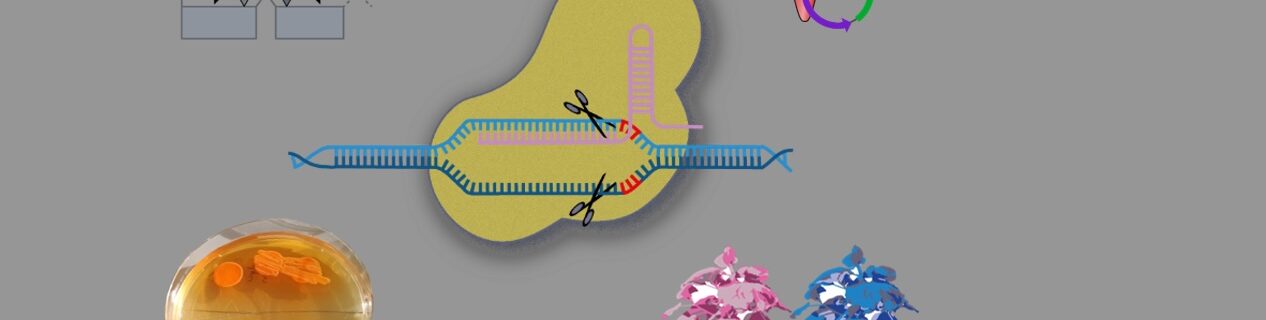
Novel technologies based on CRISPR/Cas system-derived RNA-guided nucleases have rapidly expanded during the past years, including genome editing based on nuclease-induced double strand breaks, base editing, transcriptional regulation, chromatin modification, or fluorescence labeling. In this course, we will guide through the most important innovations and their applications. Use of these technologies often requires the assembly of complex DNA constructs. Students will learn the principles of hierarchical DNA assembly and be trained in the design and in silico assembly of multi-component DNA constructs (plasmids). Further, we will design and produce genome editing constructs (with custom targets), and corresponding genotyping assays. We are planning a parallel hands-on training, during which steps of a basic genome editing experiment will be conducted. We will discuss pitfalls and obstacles. Especially for a practical part, we will focus on bacteria, but the course will encompass both pro- and eukaryotic kingdoms. Depending on applicant’s interests and needs, we will include genome editing in micro- and macro-algae, generally requiring particular reagent delivery strategies.
Objectives
At the end of the workshop, the PhD student shall be familiar with the portfolio of RGN-based technologies. Students will be able to design genome editing experiments, controlling for off-targets, to produce constructs for multiplex genome editing either in custom plasmids or published systems (in silico, wet lab), and to isolate and genotype desired individuals.
Students prerequisites
- Students need the agreement of their supervisor to attend the course.
- Participants should be interested in practical application of genome editing technologies: We will attempt to directly work with student’s data/genomes/challenges, if applicable.
- Knowledge of and experience with basic molecular biology techniques is required.
- A personal computer with administrative rights for software installation (at least temporarily before the course) is required.
Sunday 31st, March 2024 09:00








